Mass Spectrometry-based Mycology Tests Put to Use in C. auris Outbreak
A relatively new disease with particular strength in the New York City area but also in 12 U.S. states has seen 617 cases as of time of writing – with particular strength in weakened patients and those in long-term facilities. Because the fungus is detectable only with complex lab tests such as mass spectrometry, sequencing and novel PCR, test makers once again have an opportunity to demonstrate products can handle novel pathogen threats.
The fungus — Candida auris, also referred to as C. auris — is treatable with antifungal drugs called echinocandins. However, some C. auris infections are resistant to the main types of medications. It is particularly found in hospitals and long-term care facilities. The most common symptoms of invasive Candida infection are fever and chills that don’t improve after antibiotic treatment for a suspected bacterial infection. While healthy people do not often get the fungus, the CDC is warning all in such facilities to be vigilant about washing hands after patient contact.
The fungal infection tends to target those with compromised immune systems. Because these are also patients with serious conditions and existing changes of fatality, as many as half may die when contracting the condition, making isolation and treatment early on a key.
[Note – Kalorama covers mycology testing market sizing and forecasting in its report Infectious Disease Diagnostics, World Market Analysis ]
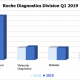
The disease has taken a particular statistical bent in the New York City area, where two-thirds of the cases were reported.
The fungal infection is tricky to diagnose, though that opens up an opportunity for mass spectrometry-based IVD systems to contribute to detection during the outbreak.
“Diagnostic devices based on matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) can differentiate C. auris from other Candida species, but not all the reference databases included in MALDI-TOF devices allow for detection,” according to the CDC guidance.
Sequencing-based molecular methods based on sequencing the D1-D2 region of the 28s rDNA or the Internal Transcribed Region (ITS) of rDNA also can identify C. auris. Other products, such as PCR, can identify fungal infection absent a specific C. auris designation:
T2 Biosystems’ T2Candida® Panel identify the most serious bacterial and fungal pathogens directly from blood sample in just three to five hours, without waiting for a positive blood culture —which can take one to six or more days. The FDA-cleared and CE marked test runs on a desktop analyzer based on magnetic resonance imaging and provides results within five hours.
Thermo Fisher’s RapID YEAST PLUS System, a qualitative micromethod employing conventional and chromogenic substrates for the identification of medically important yeast, yeast-like and related organisms which have been isolated from human clinical specimens. It does not specify C. auris at time of reporting, but can signify candida infection which can suggest further testing.
The CDC also said that laboratories with capability to characterize isolates further when C. auris is suspected are encouraged to do so. Suspected isolates can also be sent to CDC’s AR Lab Network for identification and antifungal susceptibility testing. This testing is performed free of charge and may require coordination through the State Public Health Department’s Healthcare-associated Infection (HAI) Program. The CDC advises that Isolates should be submitted on slants shipped at room temperature. Futher, mycology specific media are not necessary if the laboratory does not have them. C. auris can grow on blood or chocolate agar slants.
In China, reaction to the disease is more stated though cases are also lower and not likely to cause an epidemic in China, according to an article in in China Daily http://www.ecns.cn/news/2019-04-12/detail-ifzhhxra9209972.shtml. The country’s medical authorities are concentrating on patients with serious disease or undergoing chemotherapy.